Whole genome resequencing (WGR) is a genomic technology employed to sequence the entire genome of individual organisms or populations based on known genomic sequences. This technique facilitates a comprehensive analysis of genetic variations at both individual and population levels. Its application has notably advanced the field of breeding research, offering a rapid and efficient approach to the enhancement of both plant and animal species. This article elaborates on how WGR has accelerated the breeding process and its impact on various aspects of breeding development.
Overview of Whole Genome Resequencing
Whole genome resequencing involves sequencing an organism's entire genome to identify genetic variations compared to a reference genome. This methodology is instrumental in detecting single nucleotide polymorphisms (SNPs), insertions, deletions, and structural variants. By comparing resequenced genomes to a reference, researchers can uncover genetic differences that influence traits and contribute to genetic diversity.
Identification and Analysis of Genetic Variation Using Whole Genome Resequencing
WGR is a pivotal technique in genomics that facilitates the identification of extensive genetic variation across diverse plant populations. This comprehensive sequencing approach provides invaluable insights into the genetic diversity within and between populations, which is crucial for effective breeding and selection.
Identifying Extensive Genetic Variation
Whole genome resequencing allows for the identification of extensive genetic variation by sequencing the entire genome of multiple accessions. This approach enables the detection of a wide range of genetic variants, including SNPs, insertions, deletions, and structural variants. By analyzing these genetic variations, researchers can gain a detailed understanding of the genetic diversity present within a population and between different populations.
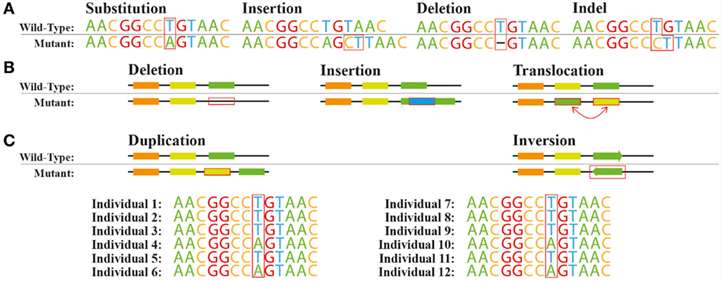
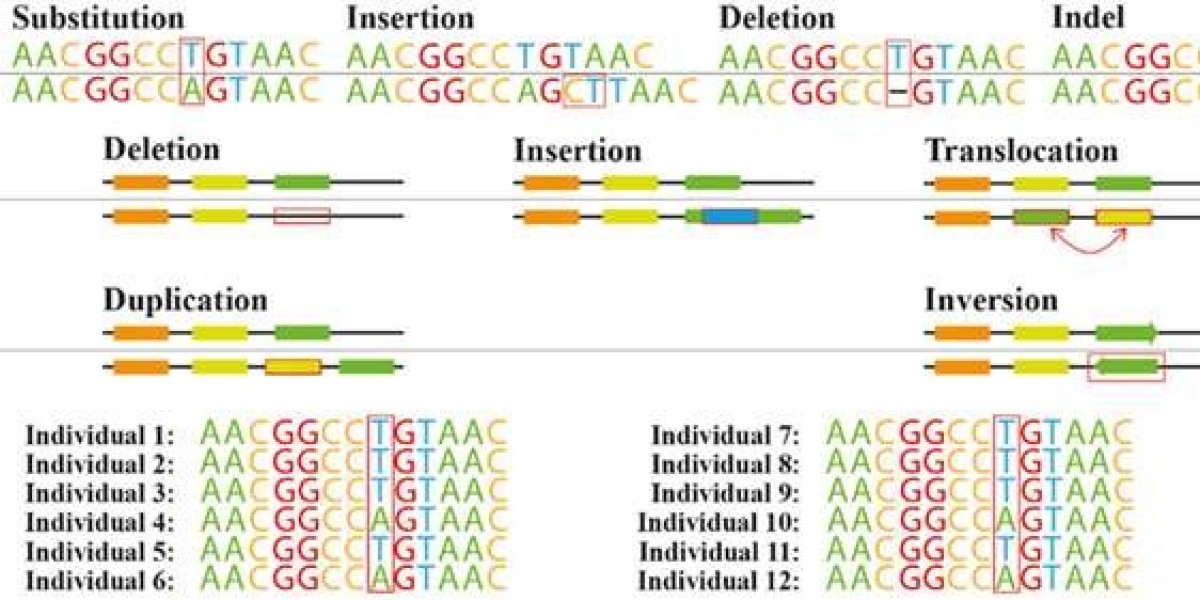
Figure 1. Common genetic variations. (João G. R. Cardoso et al,. 2015)
Detection of Genetic Variants
The capacity to identify genetic variations, including SNPs, insertions, deletions, and structural variants, represents a significant strength of WGR. SNPs, characterized by single base pair alterations within the genome, constitute the most prevalent form of genetic diversity. Insertions and deletions entail the integration or excision of nucleotide sequences, potentially affecting gene function and contributing to phenotypic variation. Structural variants, encompassing copy number variations (CNVs) among other large-scale genomic alterations, can modulate gene expression and are implicated in complex traits.
Application in Breeding Programs
The extensive genetic data acquired through WGR plays a pivotal role in breeding programs. By elucidating genetic diversity both within and between populations, breeders can strategically select parent lines that possess desirable traits, thereby facilitating the development of new varieties with enhanced characteristics. For example, the identification of genetic variants linked to traits such as yield, disease resistance, and stress tolerance enables the precise selection of breeding lines that are more likely to express these advantageous traits.
Variant Detection in Second and Third Generation Whole Genome Resequencing
The advancements in WGR technologies have significantly enhanced the detection of genetic variants, particularly through the use of second and third generation sequencing methods. These technologies offer improved resolution and sensitivity for identifying a diverse array of genetic variations, including single nucleotide polymorphisms (SNPs), insertions, deletions, and structural variants. This section discusses the contributions of these technologies to variant detection, supported by examples from academic literature.
Next-Generation Sequencing
Second-generation sequencing, widely known as next-generation sequencing (NGS), has markedly transformed genomic research by offering high-throughput sequencing capabilities. This advancement facilitates the rapid and cost-effective sequencing of a vast number of genomes, thereby enabling the identification of a multitude of genetic variants.
A notable example of the impact of second generation sequencing is the work by Lander et al. (2001), which demonstrated the utility of NGS in the detection of SNPs and small insertions and deletions in the human genome. The study utilized NGS to analyze the genetic variation in a cohort of individuals, identifying thousands of SNPs and InDels that contributed to our understanding of human genetic diversity. The authors emphasized the ability of NGS to uncover previously unobserved genetic variants, providing insights into genetic diseases and population genetics.
Another significant study is by van Dijk et al. (2014), which applied NGS to plant genomics. The researchers used NGS to explore genetic variation in Arabidopsis thaliana, identifying numerous SNPs and small structural variants. The high-throughput nature of NGS enabled the comprehensive analysis of genetic diversity within the species, facilitating the identification of genes associated with important traits such as stress tolerance and disease resistance.
Long-read Sequencing
Third-generation sequencing technologies, exemplified by long-read sequencing platforms, confer several advantages over second-generation sequencing methods. Notably, these advanced technologies produce longer read lengths, thereby enhancing the resolution of complex genomic regions and the detection of structural variants.
The study by Goodwin et al. (2015) highlights the benefits of third generation sequencing in variant detection. The authors utilized long-read sequencing to analyze the genome of a human subject, revealing structural variants and complex genomic rearrangements that were challenging to detect with second generation technologies. The ability to sequence longer DNA fragments provided a more accurate representation of the genome, enhancing the detection of large-scale genetic variations.
In a plant genomics context, the work by Chin et al. (2016) demonstrated the application of third generation sequencing in the analysis of the maize genome. The researchers employed long-read sequencing to identify structural variants and large insertions and deletions, which are often missed by second generation sequencing methods. The study underscored the importance of long-read sequencing in providing a more complete and accurate view of genetic variation in crop species.
Establishing Genetic Polymorphism Databases
The genetic polymorphism data generated through second and third-generation sequencing technologies are essential for creating comprehensive genetic databases. These databases serve as a foundation for further research, including the investigation of evolutionary relationships and the identification of candidate genes.
A notable example is the study by Weigel et al. (2016), who established a genetic polymorphism database using NGS data from Arabidopsis thaliana. The researchers created a detailed database of genetic variations, including SNPs, insertions/deletions (InDels), and structural variants. This comprehensive resource has been pivotal in advancing our understanding of Arabidopsis genetics, enabling comparative genomics studies and the identification of genes associated with various traits such as disease resistance and stress tolerance.
Second- and third-generation whole genome resequencing technologies have substantially propelled the field of genetic research forward by refining the detection of genetic variants. The high-throughput nature of second-generation sequencing, combined with the long-read capabilities of third-generation platforms, provides an exhaustive view of genetic variation. This comprehensive insight is instrumental in the development of genetic databases and the enhancement of breeding programs. The given examples underscore the profound impact of these technologies on variant detection and their significant contributions to our understanding of genetic diversity.