The study of complex biological systems has a great development with the advent of proteomics, which provides a comprehensive view of the proteins present in a given sample. Mass spectrometry-based proteomics has, in particular, emerged as a crucial tool for protein identification and quantification, but the conventional data-dependent acquisition (DDA) methods that it relies on have their own set of limitations when it comes to reproducibility and proteome coverage.
Data-independent acquisition (DIA), an alternative approach, has been gaining traction as a highly promising solution to these problems. DIA, with its intriguing and novel set of principles and applications, has been revolutionizing the field of proteomics. Technical advancements in DIA have made it a highly attractive alternative to DDA, given the various advantages it offers, along with some limitations that need to be taken into account when considering its use.
This article sets out to provide a comprehensive and in-depth exploration of DIA and its principles, highlighting its potential applications in proteomics.
Principle of Data-Independent Acquisition
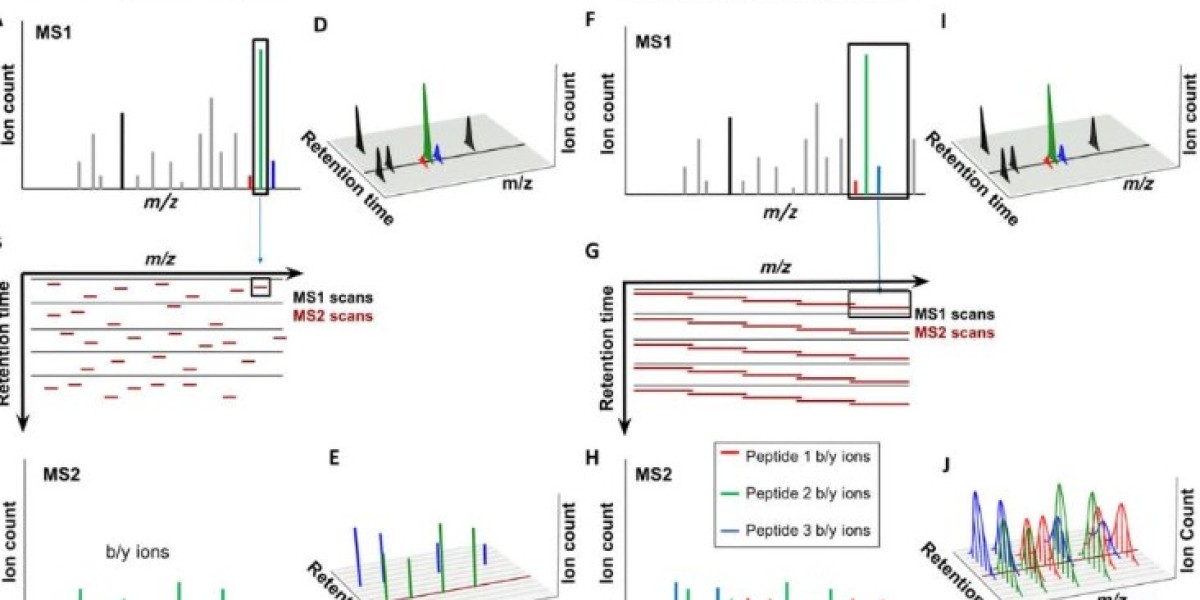
DIA's unique methodology revolves around the non-selective fragmentation of all precursor ions that fall within a pre-selected m/z range. This fragmentation process generates a series of fragment ion spectra, which are collectively known as "swaths." The real magic of DIA happens when these swaths are matched against a spectral library or spectral reference map for protein identification and quantification.
The power of DIA lies in its ability to detect and quantify an incredible number of proteins within a single sample, making it the go-to technique for large-scale proteomic studies. The fragmentation of precursor ions in a non-selective manner allows DIA to capture a vast swath of data, providing researchers with a comprehensive view of the proteome. This is in stark contrast to traditional data-dependent acquisition (DDA) methods, which tend to focus on a smaller subset of proteins and are more pro ne to errors and inconsistencies. With DIA, researchers can gettthe most complete picture of their samples possible.
Selection of precursor ions
The first step in DIA is the selection of precursor ions. This can be done in a number of ways, including by selecting a specific m/z range for fragmentation or by fragmenting all ions within a given m/z range. The latter method, known as "sequential window acquisition of all theoretical fragment-ion spectra" (SWATH), is particularly popular and has been shown to provide more comprehensive protein coverage than other methods.
Fragmentation of precursor ions
Once the precursor ions have been selected, they are fragmented using collision-induced dissociation (CID) or higher-energy collisional dissociation (HCD). The resulting fragments are then separated by mass-to-charge ratio (m/z) and detected by the MS detector. This process generates a series of fragment ion spectra, or "swaths", which represent the entire proteome within the selected m/z range.
Spectral library or spectral reference map matching
The final step in DIA is the matching of the fragment ion spectra to a spectral library or spectral reference map. A spectral library is a collection of pre-existing fragment ion spectra from known peptides, while a spectral reference map is generated by the same instrument and includes all the detectable peptides in the sample. These spectral references are used for protein identification and quantification.
Advantages OF DIA
DIA has several advantages over other MS-based proteomic techniques. First, it allows for the detection and quantification of thousands of proteins within a single sample, providing greater depth of coverage and reducing missing data. Second, it is highly reproducible, allowing for accurate and reliable protein quantification across multiple samples. Finally, DIA is compatible with a wide range of sample types, including tissue, blood, and urine, making it a versatile technique for large-scale proteomic studies.
Applications of Data-Independent Acquisition
1. Comprehensive proteome profiling
DIA-MS is an ideal technique for comprehensive proteome profiling because it enables the identification and quantification of thousands of proteins in a single experiment. DIA-MS generates data in untargeted, unbiased manner, and has been used to identify and quantify proteins across a wide range of organisms and tissues. This approach is especially useful when investigating complex biological systems where it is difficult to predict which proteins may be important. For example, a study used DIA-MS to extensively characterize the profile of more than 4000 proteins in immortalized human fetal astrocytes under distinct inflammatory conditions induced by TNF, IL-1β, and LPS, while multiplex immunoassay-based screening was used to quantify a wide range of cytokines released under these inflammatory conditions. The present study provides comprehensive information about the proteomic phenotypes of human astrocytes upon exposure to inflammatory stimuli both in monoculture and in co-culture with human brain microvascular endothelial cells.
2. Biomarker discovery
DIA-MS can be used for the discovery of novel protein biomarkers, which can be used for early disease detection, diagnosis, and treatment. DIA-MS enables the detection of low abundance proteins, which may be indicative of disease states, and allows for the measurement of protein expression levels in different biological samples. For instance, a study used DIA-MS to identify potential biomarkers that can distinguish cancer tissues from adjacent normal tissues should be developed to diagnose this disease in early stages and conduct a reliable prognostic evaluation. The proposed DIA workflow yields optimum time efficiency and data reliability and provides a good choice for proteomic analysis in biological and clinical studies, and these dysregulated proteins might be potential biomarkers for ccRCC diagnosis.cancer.
3. Drug discovery
The use of data-independent acquisition mass spectrometry (DIA-MS) has proven to be a promising technique for the identification and quantification of proteomic alterations induced by pharmaceutical agents. Through the employment of this technique, we may glean valuable insights into the underlying mechanisms governing drug efficacy, a paramount consideration in the pursuit of drug discovery and development.
In a recent study, a team of intrepid researchers sought to construct a comprehensive DIA spectrum library of the proteome of pancreatic cancer, thereby allowing for the analysis of proteomic changes in pancreatic cells before and after treatment with the drug gemcitabine (EGM). This approach facilitated the elucidation of the GEM-sensitive and epithelial-to-mesenchymal transition (EMT) phenotypes of the pancreatic cell lines, thereby presenting a rich source of information for the deepening of proteomic analyses of pancreatic diseases. The findings of this research hold tremendous potential for the development of more targeted and efficacious therapeutics, thus ushering in a new era of safe and effective treatments for patients.
4. Systems biology
With its remarkable sensitivity and precision, DIA-MS is proving to be an exceptional tool in the increasingly vital field of systems biology studies. DIA-MS's ability to identify and quantify proteins provides invaluable insights into protein-protein interactions, signaling pathways, and cellular processes, and cracking open new possibilities for medical and biotechnological breakthroughs. In addition, DIA-MS can be paired with other omics technologies, such as genomics and transcriptomics, providing a broader and more comprehensive understanding of biological systems. This potent combination of technologies enhances the versatility of DIA-MS, leading to a deeper understanding of the intricate connections and complexities of biological systems.
For example, the researchers generated a spectral assay library for 4,014 of the 4,389 annotated E. coli proteins using one- and two-dimensional fractionated samples, and ion mobility separation enabling deep proteome coverage. They demonstrate the utility of this high-quality library with robustness in quantitation of the E. coli proteome and with rapid-chromatography to enhance throughput by targeted DIA-MS.The results provided insights into the metabolic pathways and cellular processes that are important for the survival of the bacterium.
References:
- Dozio, V., Sanchez, JC. Profiling the proteomic inflammatory state of human astrocytes using DIA mass spectrometry. J Neuroinflammation 15, 331 (2018)
- Yimeng Song et al,. Data-Independent Acquisition-Based Quantitative Proteomic Analysis Reveals Potential Biomarkers of Kidney Cancer. Proeomics clinical applications(2017).
- Kong, R., Qian, X. & Ying, W. Pancreatic cancer cells spectral library by DIA-MS and the phenotype analysis of gemcitabine sensitivity. Sci Data 9, 283 (2022).